Math-Physics-ML MCP System
GPU-accelerated Model Context Protocol servers for computational mathematics, physics simulations, and machine learning.
📚 Documentation
| Guide | Description |
|---|---|
| Installation | Setup instructions for pip, uv, and uvx |
| Configuration | Claude Desktop & Claude Code setup |
| Quick Start | Get running in 5 minutes |
| API Reference | Complete tool documentation |
| Visual Demos | Interactive physics simulations |
About
This system enables AI assistants to perform real scientific computing — from solving differential equations to running molecular dynamics simulations.
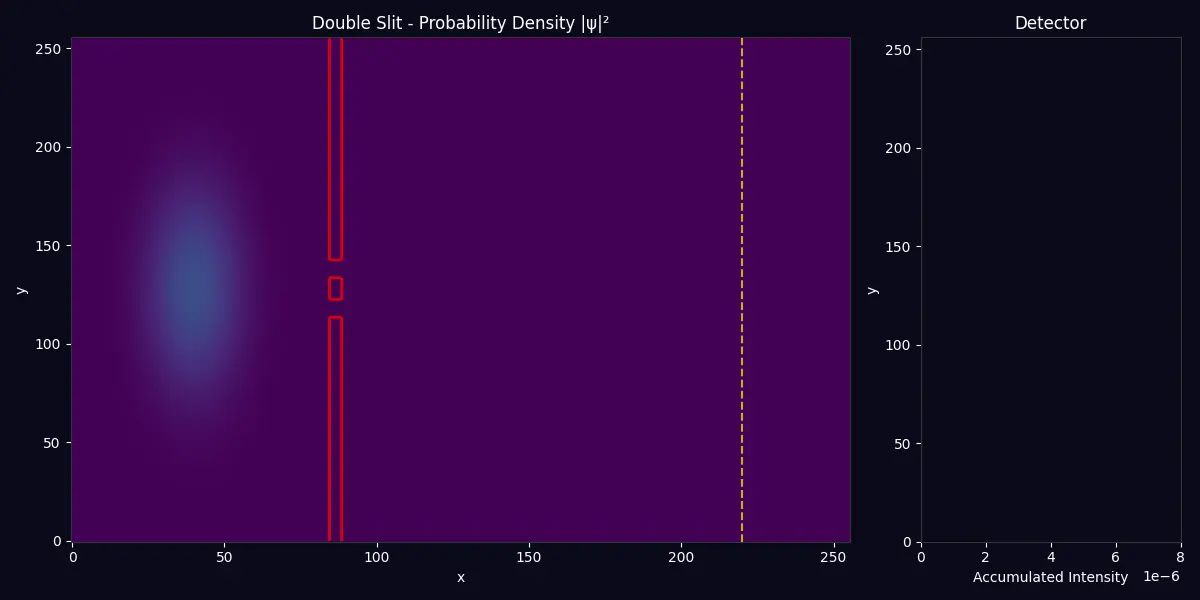
Quantum Wave Mechanics Double-slit interference pattern from solving the time-dependent Schrödinger equation |
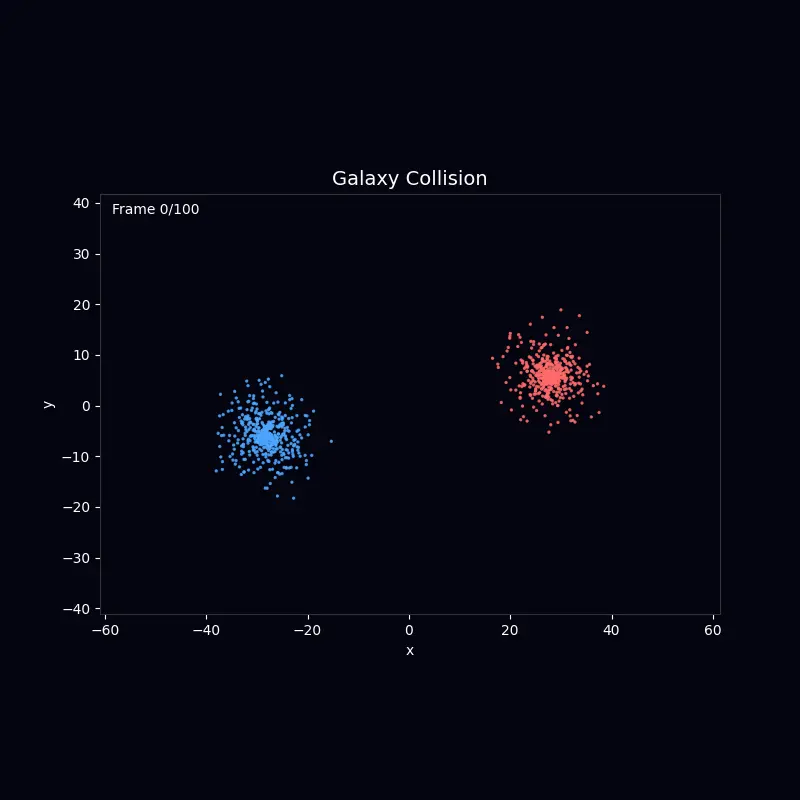
N-Body Dynamics Galaxy merger simulation using gravitational N-body calculations |
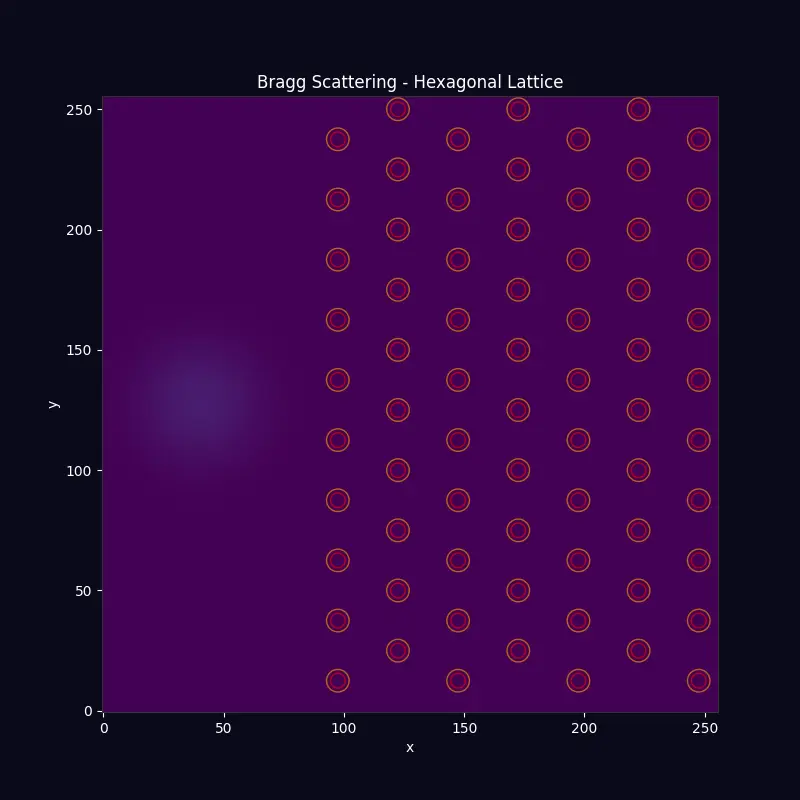
Crystal Diffraction Bragg scattering from a hexagonal (graphene-like) lattice |
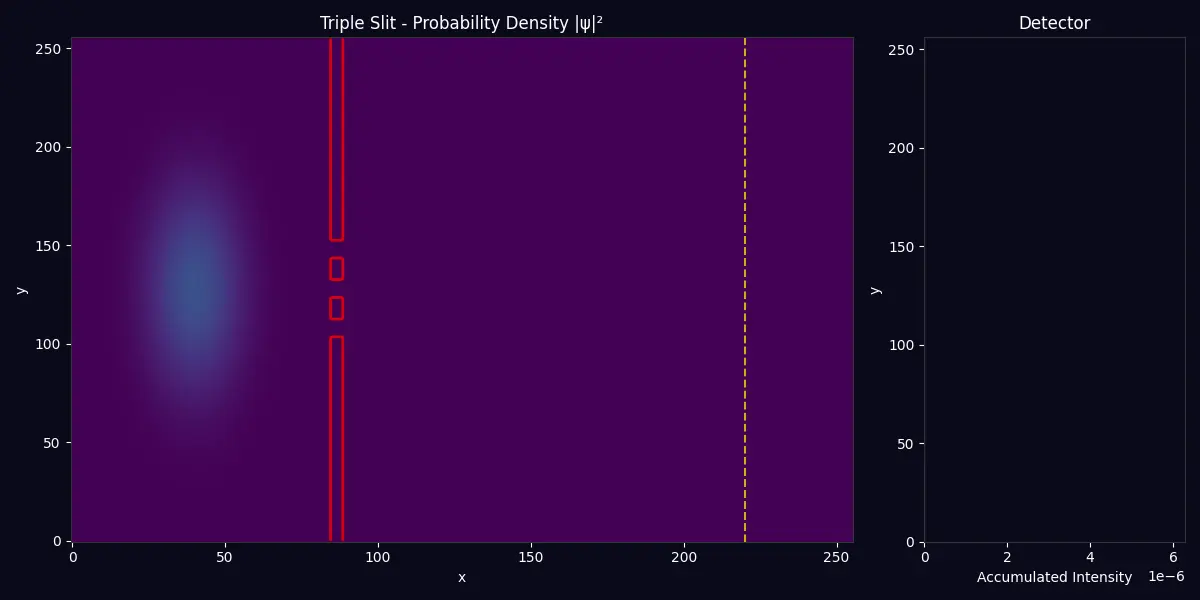
Multi-Slit Interference Complex interference patterns from three coherent sources |
Overview
This system provides 4 specialized MCP servers that bring scientific computing capabilities to AI assistants like Claude:
| Server | Description | Tools |
|---|---|---|
| Math MCP | Symbolic algebra (SymPy) + numerical computing | 14 |
| Quantum MCP | Wave mechanics & Schrodinger simulations | 12 |
| Molecular MCP | Classical molecular dynamics | 15 |
| Neural MCP | Neural network training & evaluation | 16 |
Key Features:
- GPU acceleration with automatic CUDA detection (10-100x speedup)
- Async task support for long-running simulations
- Cross-MCP workflows via URI-based data sharing
- Progressive discovery for efficient tool exploration
Quick Start
Installation with uvx (Recommended)
Run any MCP server directly without installation:
# Run individual servers
uvx scicomp-math-mcp
uvx scicomp-quantum-mcp
uvx scicomp-molecular-mcp
uvx scicomp-neural-mcp
Installation with pip/uv
# Install individual servers
pip install scicomp-math-mcp
pip install scicomp-quantum-mcp
pip install scicomp-molecular-mcp
pip install scicomp-neural-mcp
# Or install all at once
pip install scicomp-math-mcp scicomp-quantum-mcp scicomp-molecular-mcp scicomp-neural-mcp
# With GPU support (requires CUDA)
pip install scicomp-math-mcp[gpu] scicomp-quantum-mcp[gpu] scicomp-molecular-mcp[gpu] scicomp-neural-mcp[gpu]
Configuration
Claude Desktop
Add to your Claude Desktop configuration file:
macOS: ~/Library/Application Support/Claude/claude_desktop_config.json
Windows: %APPDATA%\Claude\claude_desktop_config.json
{
"mcpServers": {
"math-mcp": {
"command": "uvx",
"args": ["scicomp-math-mcp"]
},
"quantum-mcp": {
"command": "uvx",
"args": ["scicomp-quantum-mcp"]
},
"molecular-mcp": {
"command": "uvx",
"args": ["scicomp-molecular-mcp"]
},
"neural-mcp": {
"command": "uvx",
"args": ["scicomp-neural-mcp"]
}
}
}
Claude Code
Add to your project's .mcp.json:
{
"mcpServers": {
"math-mcp": {
"command": "uvx",
"args": ["scicomp-math-mcp"]
},
"quantum-mcp": {
"command": "uvx",
"args": ["scicomp-quantum-mcp"]
}
}
}
Or configure globally in ~/.claude/settings.json.
Usage Examples
Math MCP
# Solve equations symbolically
symbolic_solve(equations="x**3 - 6*x**2 + 11*x - 6")
# Result: [1, 2, 3]
# Compute derivatives
symbolic_diff(expression="sin(x)*exp(-x**2)", variable="x")
# Result: cos(x)*exp(-x**2) - 2*x*sin(x)*exp(-x**2)
# GPU-accelerated matrix operations
result = matrix_multiply(a=matrix_a, b=matrix_b, use_gpu=True)
Quantum MCP
# Create a Gaussian wave packet
psi = create_gaussian_wavepacket(
grid_size=[256],
position=[64],
momentum=[2.0],
width=5.0
)
# Solve time-dependent Schrodinger equation
simulation = solve_schrodinger(
potential=barrier_potential,
initial_state=psi,
time_steps=1000,
dt=0.1,
use_gpu=True
)
Molecular MCP
# Create particle system
system = create_particles(
n_particles=1000,
box_size=[20, 20, 20],
temperature=1.5
)
# Add Lennard-Jones potential
add_potential(system_id=system, potential_type="lennard_jones")
# Run MD simulation
trajectory = run_nvt(system_id=system, n_steps=100000, temperature=1.0)
# Analyze diffusion
msd = compute_msd(trajectory_id=trajectory)
Neural MCP
# Define model
model = define_model(architecture="resnet18", num_classes=10, pretrained=True)
# Load dataset
dataset = load_dataset(dataset_name="CIFAR10", split="train")
# Train
experiment = train_model(
model_id=model,
dataset_id=dataset,
epochs=50,
batch_size=128,
use_gpu=True
)
# Export for deployment
export_model(model_id=model, format="onnx", output_path="model.onnx")
Development
# Clone the repository
git clone https://github.com/andylbrummer/math-mcp.git
cd math-mcp
# Install dependencies
uv sync --all-extras
# Install MCP servers in editable mode (required for entry points)
uv pip install --python .venv/bin/python \
-e servers/math-mcp \
-e servers/quantum-mcp \
-e servers/molecular-mcp \
-e servers/neural-mcp
# Run tests
uv run pytest -m "not gpu" # CPU only
uv run pytest # All tests (requires CUDA)
# Run with coverage
uv run pytest --cov=shared --cov=servers
Note: The editable install step is required because
uv syncdoesn't install entry point scripts for workspace packages. After this step, you can run servers directly withuv run scicomp-math-mcp.
See CONTRIBUTING.md for development guidelines.
Performance
GPU acceleration provides significant speedups for compute-intensive operations:
| MCP | Operation | CPU | GPU | Speedup |
|---|---|---|---|---|
| Math | Matrix multiply (4096x4096) | 2.1s | 35ms | 60x |
| Quantum | 2D Schrodinger (512x512, 1000 steps) | 2h | 2min | 60x |
| Molecular | MD (100k particles, 10k steps) | 1h | 30s | 120x |
| Neural | ResNet18 training (1 epoch) | 45min | 30s | 90x |
Architecture
For technical details about the system architecture, see ARCHITECTURE.md.
License
MIT License - see LICENSE for details.
Contributing
Contributions are welcome! Please see CONTRIBUTING.md for guidelines.





