MCP Proto-OKN Server
A Model Context Protocol (MCP) server providing seamless access to SPARQL endpoints with specialized support for the NSF-funded Proto-OKN Project (Prototype Open Knowledge Network). This server enables querying the scientific knowledge graphs hosted on the FRINK platform. In addition, third-party SPARQL endpoints can be queried.
Features
- 🔗 FRINK Integration: Automatic detection and documentation linking for FRINK-hosted knowledge graphs
- 🕸️ Proto-OKN Knowledge Graphs: Optimized support for biomedical and scientific knowledge graphs, including:
- 🧬 Biology & Health
- 🌱 Environment
- ⚖️ Justice
- 🛠️ Technology & Manufacturing
- ⚙️ Flexible Configuration: Support for both FRINK and custom SPARQL endpoints
- 📚 Automatic Documentation: Registry links and metadata for Proto-OKN knowledge graphs
- 🔗 Federated Query: Prompts can query multiple endpoints
Architecture
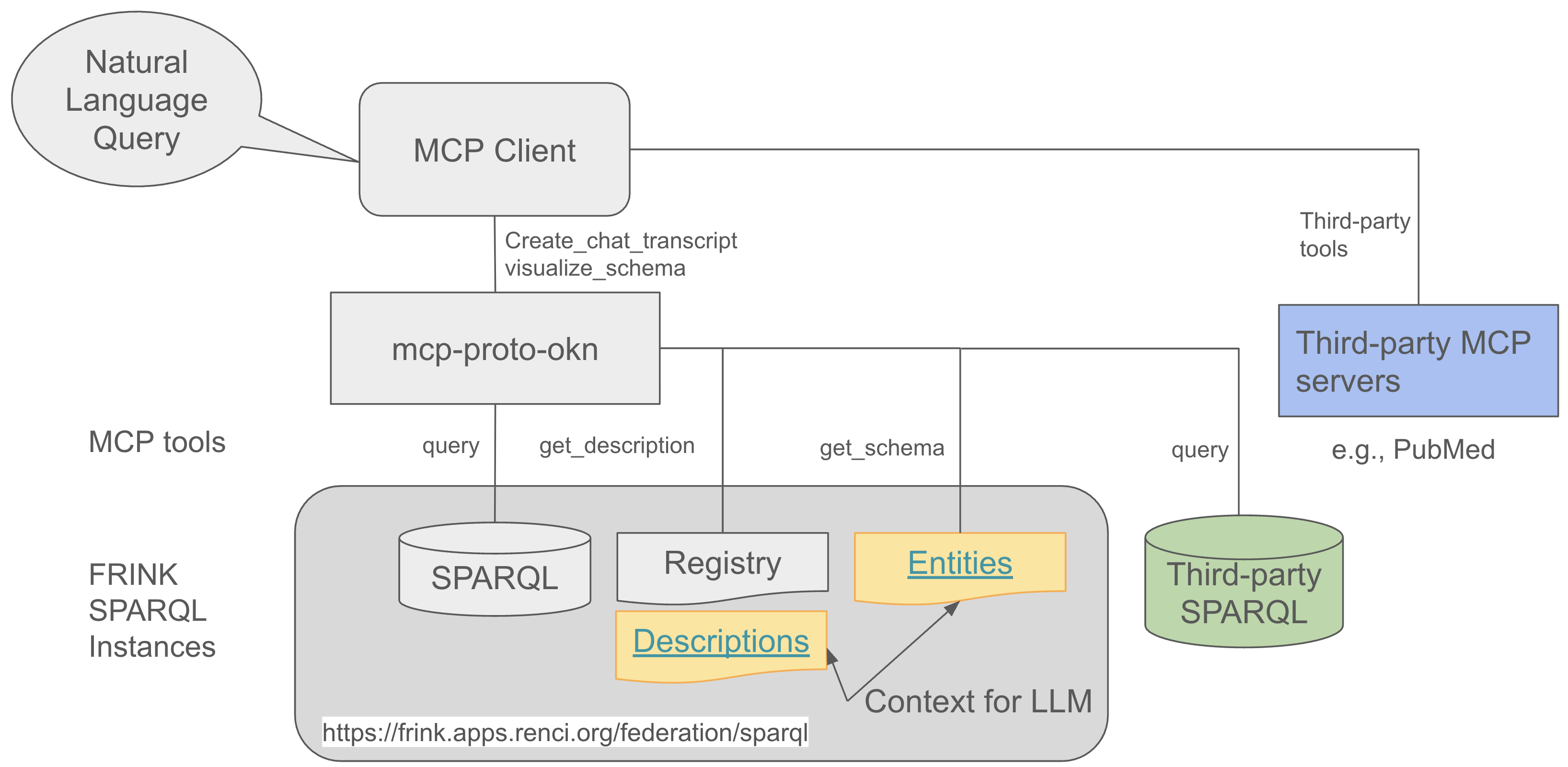
The MCP Server Proto-OKN acts as a bridge between AI assistants (like Claude) and SPARQL knowledge graphs, enabling natural language queries to be converted into structured SPARQL queries and executed against scientific databases.
Prerequisites
Before installing the MCP Server Proto-OKN, ensure you have:
- Operating System: macOS, Linux, or Windows
- Client Application: One of the following:
- Claude Desktop with Pro or Max subscription
- VS Code Insiders with GitHub Copilot subscription
Installation
Installation instructions for Claude Desktop and VS Code Insiders
Quick Start
Once configured, you can start querying knowledge graphs through natural language prompts in Claude Desktop or VS Code chat interface.
Select and Configure MCP Tools (Claude Desktop)
From the top menu bar:
1. Select: Claude->Settings->Connectors
2. Click: Configure for the MCP endpoints you want to use
3. Select Tool permissions: Always allow
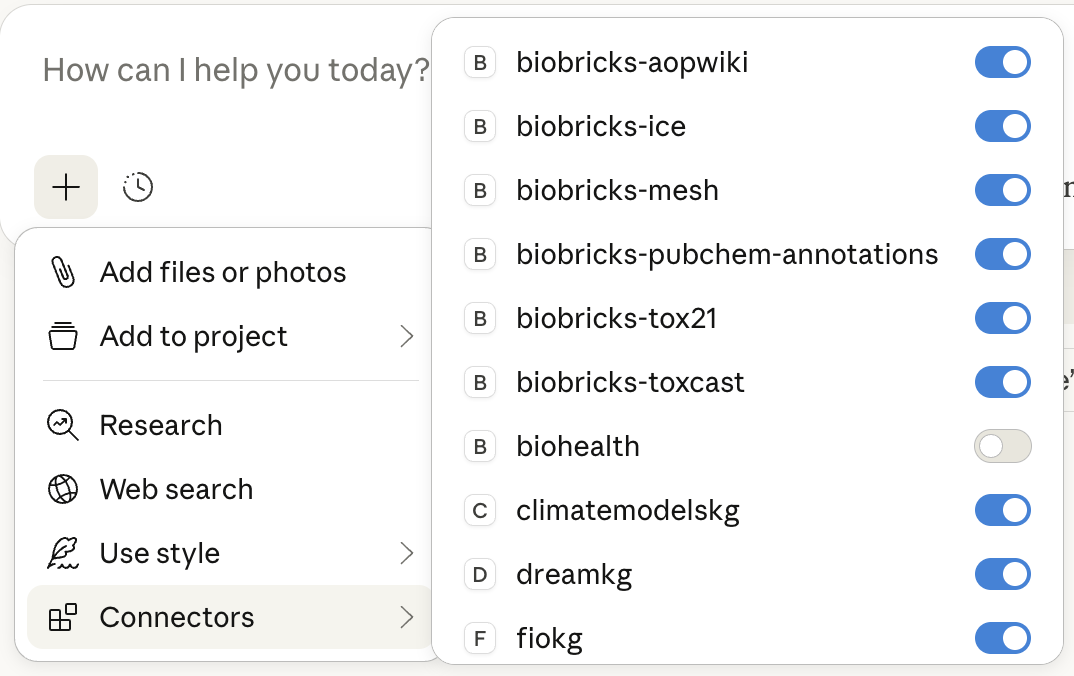
In the prompt dialog box, click the + button:
1. Turn off Web search
2. Toggle MCP services on/off as needed
Use @kg_name to refer to a specific knowledge graph in chat (for example, @spoke-genelab).
To create a transcript of a chat (see examples below), use the following prompt:
Create a chat transcript.
The transcript can then be downloaded in .md or .pdf format.
Example Queries
Knowledge Graph Overviews & Class Diagrams
Each link below points to a chat transcript that demonstrates how to generate a knowledge-graph overview and class diagram for a given Proto-OKN Theme 1 KG. The examples are grouped by domain area.
| 🧬 Biology & Health | 🌱 Environment | ⚖️ Justice | 🛠️ Technology & Manufacturing | Other |
|---|---|---|---|---|
| biobricks-aopwiki | sawgraph | ruralkg | securechainkg | nasa-gesdisc-kg |
| biobricks-ice | fiokg | scales | sudokn | |
| biobricks-mesh | geoconnex | nikg | ||
| biobricks-pubchem-annotations | spatialkg | dreamkg | ||
| biobricks-tox21 | hydrologykg | |||
| biobricks-toxcast | ufokn | |||
| spoke-genelab | wildlifekn | |||
| spoke-okn | climatemodelskg | |||
| sockg |
Use Cases
-
Spaceflight Gene Expression Analysis (spoke-genelab, spoke-okn)
-
Spaceflight Gene Expression with Literature Analysis (spoke-genelab, spoke-okn, PubMed)
-
Disease Prevalence - Socio-Economic Factors Correlation (spoke-okn)
-
Environmental Justice (sawgraph, scales, spatialkg, spoke-okn)
-
Flooding and Socio-Economic Factors (ufokn, spatialkg, spoke-okn)
Proto-OKN Integration Opportunities
Cross-Platform LLM Benchmarks
This section compares the results of two queries using Claude Desktop and VS Code Insiders with commons LLMs.
| Query | Claude Sonnet 4.5 | Claude Sonnet 4.5 | Gemini 3 Pro | Groq Code Fast 1 | GPT-5.2 |
|---|---|---|---|---|---|
| Spaceflight Missions | Claude Desktop | VS Code | VS Code | VS Code | VS Code |
| Gene Expression Analysis | Claude Desktop | VS Code | VS Code | VS Code | VS Code |
Building and Publishing (maintainers only)
Instructions for building, testing, and publishing the mcp-proto-okn package on PyPI
API Reference and Query Analysis System
Troubleshooting
MCP server not appearing in Claude Desktop:
- Ensure you've completely quit and restarted Claude Desktop (not just closed the window)
- Check that your JSON configuration is valid (attach your config file to a chat and ask it to fix any errors)
- Verify that
uvxis installed and accessible in your PATH (which uvx)
Connection errors:
- Verify the SPARQL endpoint URL is correct and accessible
- Some endpoints may have rate limits or temporary downtime
Performance issues:
- Complex SPARQL queries may take time to execute
- Consider breaking down complex queries into smaller parts
License
This project is licensed under the BSD 3-Clause License. See the LICENSE file for details.
Citation
If you use MCP Server Proto-OKN in your research, please cite the following works:
@software{rose2025mcp-proto-okn,
title={MCP Server Proto-OKN},
author={Rose, P.W. and Nelson, C.A. and Saravia-Butler, A.M. and Shi, Y. and Baranzini, S.E.},
year={2025},
url={https://github.com/sbl-sdsc/mcp-proto-okn}
}
@software{rose2025spoke-genelab,
title={NASA SPOKE-GeneLab Knowledge Graph},
author={Rose, P.W. and Nelson, C.A. and Gebre, S.G. and Saravia-Butler, A.M. and Soman, K. and Grigorev, K.A. and Sanders, L.M. and Costes, S.V. and Baranzini, S.E.},
year={2025},
url={https://github.com/BaranziniLab/spoke_genelab}
}
Related Publications
-
Nelson, C.A., Rose, P.W., Soman, K., Sanders, L.M., Gebre, S.G., Costes, S.V., Baranzini, S.E. (2025). "Nasa Genelab-Knowledge Graph Fabric Enables Deep Biomedical Analysis of Multi-Omics Datasets." NASA Technical Reports, 20250000723. Link
-
Sanders, L., Costes, S., Soman, K., Rose, P., Nelson, C., Sawyer, A., Gebre, S., Baranzini, S. (2024). "Biomedical Knowledge Graph Capability for Space Biology Knowledge Gain." 45th COSPAR Scientific Assembly, July 13-21, 2024. Link
Acknowledgments
Funding
This work is in part supported by:
- National Science Foundation Award #2333819: "Proto-OKN Theme 1: Connecting Biomedical information on Earth and in Space via the SPOKE knowledge graph"
Related Projects
- Proto-OKN Project - Prototype Open Knowledge Network initiative
- FRINK Platform - Knowledge graph hosting infrastructure
- Knowledge Graph Registry - Catalog of available knowledge graphs
- Model Context Protocol - AI assistant integration standard
- Original MCP Server SPARQL - Base implementation reference
For questions, issues, or contributions, please visit our GitHub repository.



